Cell Segmentation
BY User-Trainable AI
Segmentation of cellular sub-structures is the first and crucial step in many biological image analysis applications. Current approaches usually involve complex algorithms that are difficult to make robust. Here, we demonstrate how highly accurate segmentation results can be achieved with an intuitive workflow, which turns explicit visual user input into a reliable method for quantifying cellular phenotypes.
Segmentation is important but usually cumbersome.
From High-Content screening to cell-production quality control, many high-value investigations depend on reliable segmentation of microscopic images into biologically meaningful sub-regions. Typical examples of such segmentation tasks are detection of the nucleus, the cytoplasm, or various specific structures like vesicles, neurites, and many more. There are mainly two types of microscopic images to be considered: Label-free (brightfield, phase-contrast, etc.) and fluorescently labeled (either with dyes or antibodies coupled to fluorophores). Label-free images are notoriously difficult to segment using classical approaches. With the method presented here, very accurate results can be achieved on label-free images as well. But for the sake of better comparison to the usual situation, we will focus on fluorescently labeled images in this note.
Classically, fluorescence microscopy images are segmented based on some sophisticated usage of intensity thresholding. Flatfield normalization can be used to remove the confounding effects of uneven illumination. Complex algorithms can then be utilized to "grow" cleverly identified seed-regions in a controlled fashion, performing local thresholding at each step, for example. These algorithms can be implemented by hand in various scripting languages, or they may already be implemented in software packages with a graphical user interface, in the form of building blocks. In all cases, the main task for the user of such methods is to tune the parameters of the algorithm until the desired segmentation result is achieved. This is where the main issues arise: The need for a highly specific skill set in the case of scripting languages or highly specific experience in the case of building blocks strongly limit the usefulness of these methods. Just as problematic are the abstractness of the parameters, their lack of direct biological meaning, the huge size of the search space for a viable solution and - despite the extremely high complexity - their lack of power to encompass very common variations in biological phenotypes. Very often, this results in frustrating experiences when trying to adapt multiple parameters to remove a visually obvious flaw in the segmentation that even a non-expert could easily understand and correct on each image.
Universal AI for all segmentation tasks is not practical.
It might be tempting to take advantage of the power and trainability of segmentation AI to create one highly trained algorithm that has seen many different types of training sets in the hopes of generating a universal segmentation tool. Such an algorithm could then be used without any further user input and just "give the right answer" when confronted with any microscopic image of cells. While admiring the ambition behind such an endeavor, we deem it to be unpractical. There are several reasons: Firstly, there are a huge number of cell types, imaging modalities, assay conditions, time points, etc. that have profound impact on the data the algorithm would have to be able to cope with. This makes the size and diversity of the required training set forbiddingly large. Secondly, what the right answer is, depends on the question the researcher wants to ask of the images. There could be situations where the shape of nuclei is critical or others, where a specific sub-structure that extends throughout the cell is relevant. To create one algorithm that has the answer to all possible questions is not realistic.
Segmentation is easy for humans and AI.
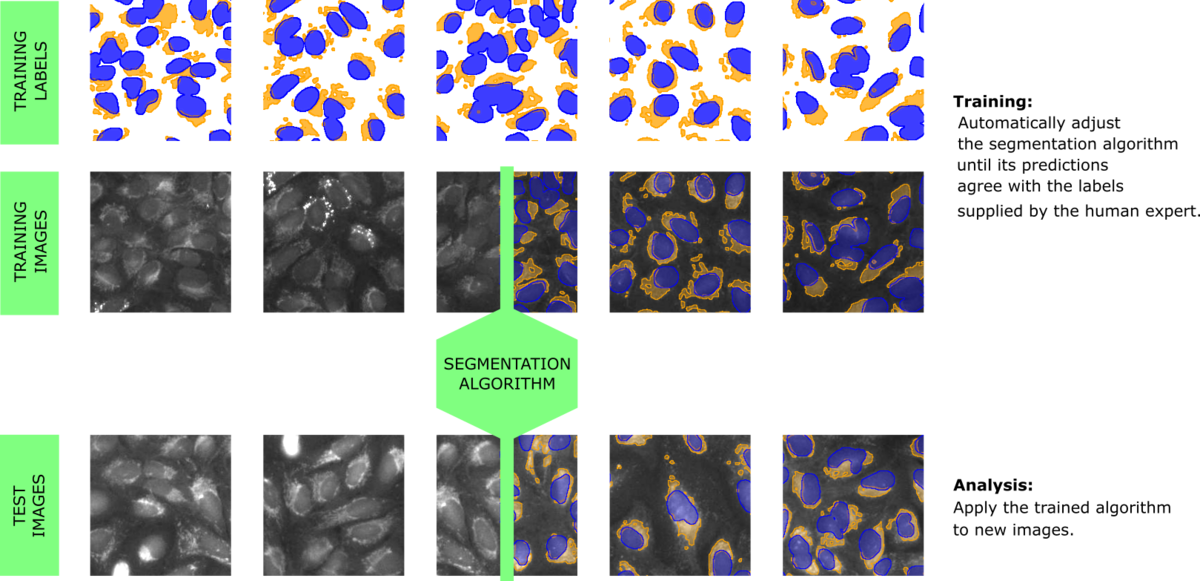
A type of algorithm that is very adept at solving segmentation tasks the way humans do is based on artificial neural networks, which takes us into the realm of deep learning / artificial intelligence, or AI for short. These algorithms are based on a computational structure that mimics a huge number of connected neurons. The way in which the neurons interact is tuned automatically, by running many examples through the network and demanding that the output of the network solves a specific task - for example a particular segmentation of the example-images (Figure 1). This provides the decisive property why such algorithms interface well with a human: The human merely needs to produce segmented images for training, which is a relatively easy task. During training, the AI is "rewarded" if it predicts the user-supplied segmentation and thus keeps improving itself while repeatedly processing the training set of images and labels.
The strength of such AI algorithms is that they develop the discerning features automatically. If a certain shape or pattern is more relevant to finding e.g. nucleoli than the raw intensity, the AI will gradually develop these useful "notions" internally, encoding them in its own set of artificial neurons. In contrast, a classical algorithm is based solely on the features that the expert designer of that algorithm found useful during development based on their specific training set. This is of course very limiting in terms of transferring the algorithm to slightly different applications.
User-trainable AI generates robust, application-specific segmentation methods.
Therefore, our approach is to create a workflow that produces algorithms trained to answer specific questions on specific types of data sets. As mentioned above, the ability to learn the right parameters for solving a specific question is a characteristic of AI-type algorithms. To take full advantage of this strength, one crucial ingredient is the interface between the human providing their biological expert knowledge and the AI, which is initially powerful but completely ignorant. The biologist needs to be able to focus on biological content and everything else must be taken care of by the software. We have developed a powerful yet simple user-interface to generate labeled images that can be used for training. One key feature is the iterative nature of the training process: After labeling a single image and pre-training the AI once, the user is presented with AI-generated proposals for segmenting additional images. The interface merely requires the user to correct the labels in those regions where the AI-proposal was not yet correct. This has the combined effect of greatly reducing the time and effort for labeling and enabling the AI to focus on examples where it needs to improve.
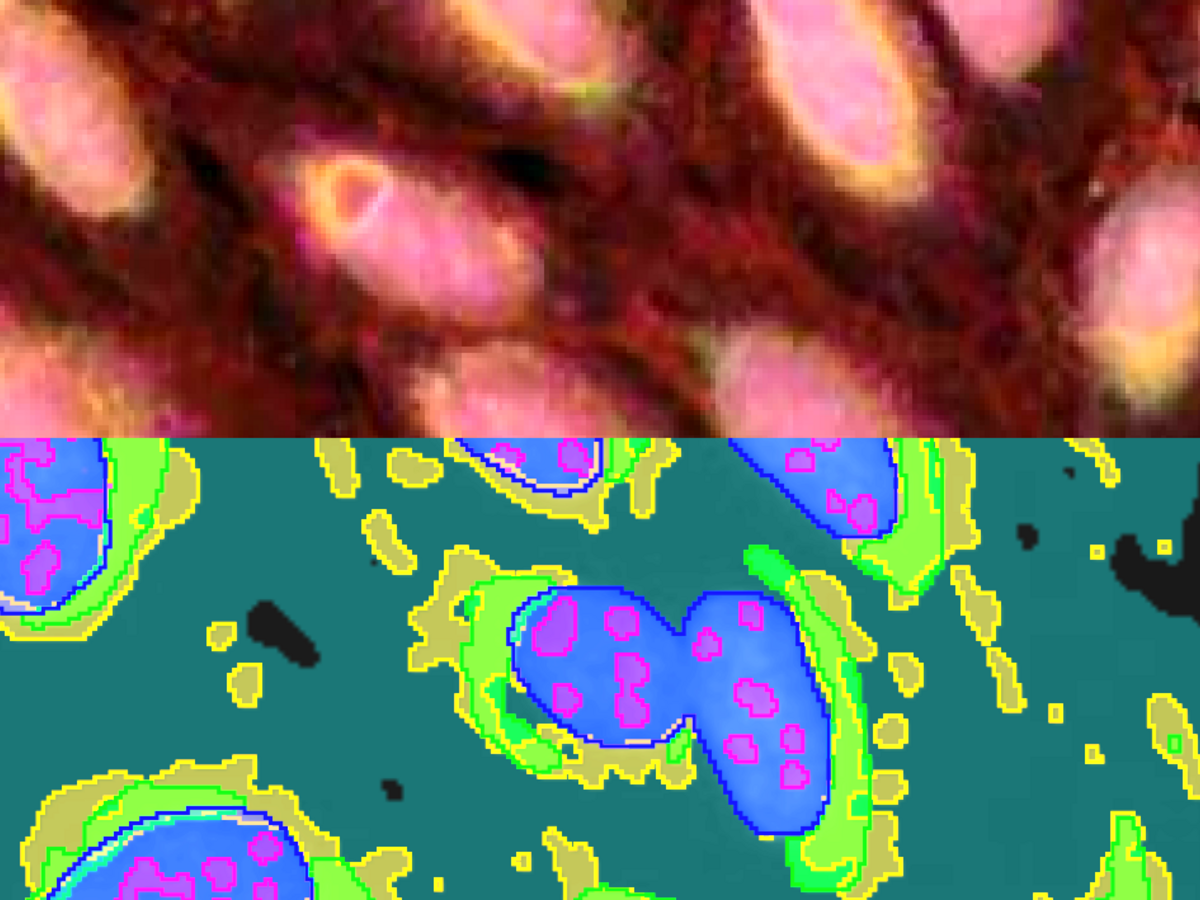
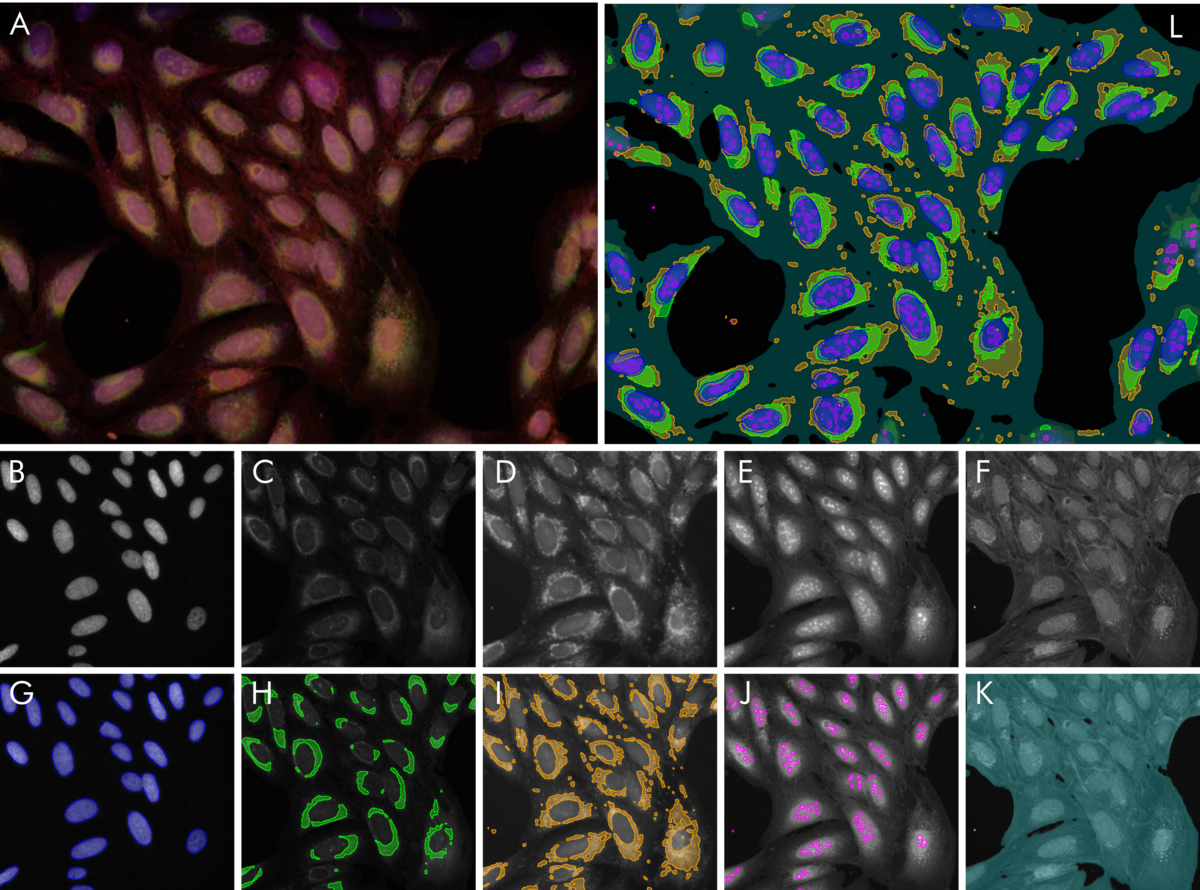
To illustrate the power of this approach, we have trained 5 algorithms to solve typical segmentation tasks on a multi-channel fluorescence dataset published by the Broad Institute (see Reference). The results are illustrated in Figure 2. Nucleus segmentation is performed after labeling based on the nuclear stain Hoechst. The results were already near perfect after a single labeled image. Nucleoli, endoplasmic reticulum, mitochondria are detected by their own algorithms, respectively. No algorithm needed more than one fully labeled image and at most one or two images which were partially corrected. In total, the effort of labeling by the user amounted to less than one hour. The outcome are five segmentation methods which can be immediately used to robustly generate quantitative results for a plethora of biological questions: Size, shape and number of nuclei and nucleoli. Relative areas and intensities of organelles like mitochondria, or total confluency. With a little post-processing, the number of useful output parameters can be increased drastically: How strong is the striation in the non-nuclear regions of the cell due to the actin stain? What is the overlap between the mitochondria region and the ER-region? And so on.
A universal workflow
The approach is so versatile that it readily translates to a huge number of applications. Firstly, it has been shown to produce excellent results on label-free images. This is an area where classical methods routinely fail, due to their predominant reliance on intensity for segmentation. However, with the user-trainable AI-approach, remarkable precision in segmenting sub-cellular structure has been achieved. Secondly, in the case of 3D structures like organoids, the trainability has been shown to be very useful. With the same small training effort as in the case of 2D cells, organoids can be segmented, and their substructure can be analyzed without the need for custom scripting.

"Our customers have challenged VAIDR with a very diverse set of scientific problems over the past three years. I have been surprised again and again, at how versatile the VAIDR workflow has proven to be. Usually, we were able to make a significant contribution to our partner's research project."
Bruno Chilian
CSO at TRI
Reference:
Gustafsdottir SM, Ljosa V, Sokolnicki KL, Anthony Wilson J, Walpita D, Kemp MM, et al. (2013) Multiplex Cytological Profiling Assay to Measure Diverse Cellular States. PLoS ONE 8(12): e80999. doi.org/10.1371/journal.pone.0080999.